
Note 2: You can optionally use the Viewing panel (side view) to alter the image as we have seen in a previous section to add a silhouette contour or change the lighting option e.g. Use the button Reverse ordering of above if necessary. Note 1: to draw a scale click the button at the very bottom Create corresponding color key and use the mouse to draw at the desired size. Keep the default values as well as the default suggested 3-colors of red, white and blue. Click Apply button o create an approximate electrostatic Colombic surface coloring.The she top part of the window will list the surface(s) currently available within Chimera. Tools > Surface/Binding Analysis > Coulombic Surface Coloring will open a window meant to color surfaces.
#UCSF CHIMERA FREE DOWNLOAD SOFTWARE#
Software that calculate a Poisson-Boltzmann grid are not included in Chimera.However, Chimera does include interfaces to such programs: DelPhiController requires a local (user-installed) copy of DelPhi, and the APBS tool can use either a web service or a locally installed copy of APBS (Adaptive Poisson-Boltzmann Solver). However, a Coulombic potential may suffice for visualization. Note: Poisson-Boltzmann calculations are more complex and, if done correctly, more accurate than simple Coulomb’s law approaches. A distance-dependent dielectric ( \(\epsilon = Cd\) where \(C\) is some constant) is sometimes used to approximate screening by implicit solvent.
#UCSF CHIMERA FREE DOWNLOAD HOW TO#
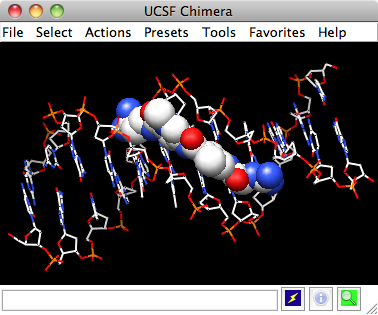
In this exercise we will explore how to show hydrogen bonds and using selection methods to show them only for specific residues, in our example on the hydroxyl of Serine 92 ( SER 92.A OC.)
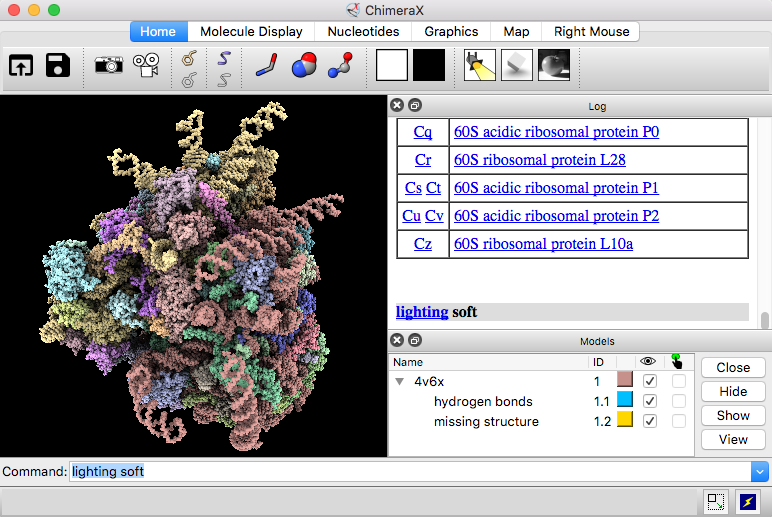
The Publication menus alter the view prepared by the Interactive menu. Note: the Interactive menu options create a view. Publication 3 (depth-cued, rounded ribbon) Publication 1 (silhouette, rounded ribbon) Protein is wireframe, ligands are spheres The difference is that the ribbon is rainbow-colored from a blue-colored N-termial blue to a red-colored C-terminus.Įngage the Presets menus to explore the preset representations that are offered. The first one ** Presets > Interactive1(ribbon)** looks very similar to the current display when a molecule is opened. Once you have found a suitable view, you can try the “ Presets” menus that alter the display of the molecule with preset options.


 0 kommentar(er)
0 kommentar(er)
